Creating a Population#
My Drive/GitHub/transmission_ms/coalescence_time.ipynb
Checked in working order 6 Nov 2022
# Create a path to \\GitHub\\transmission_ms
# .. which contains the definitive version of coalestr.py
# .. this is not necessary when working within \\GitHub\\transmission_ms
import sys
sys.path.insert(0, 'C:\\Users\\dominic\\H_Drive\\My Drive\\GitHub\\transmission_ms')
import coalestr as cs
import matplotlib.pyplot as plt
N = 300 # Effective number of hosts
Q = 5 # Quantum of transmission
X = 0.5 # Crossing rate of transmission chains
duration = 50000
history = [[duration, N, Q, X, 0]]
my_village = cs.Population(history)
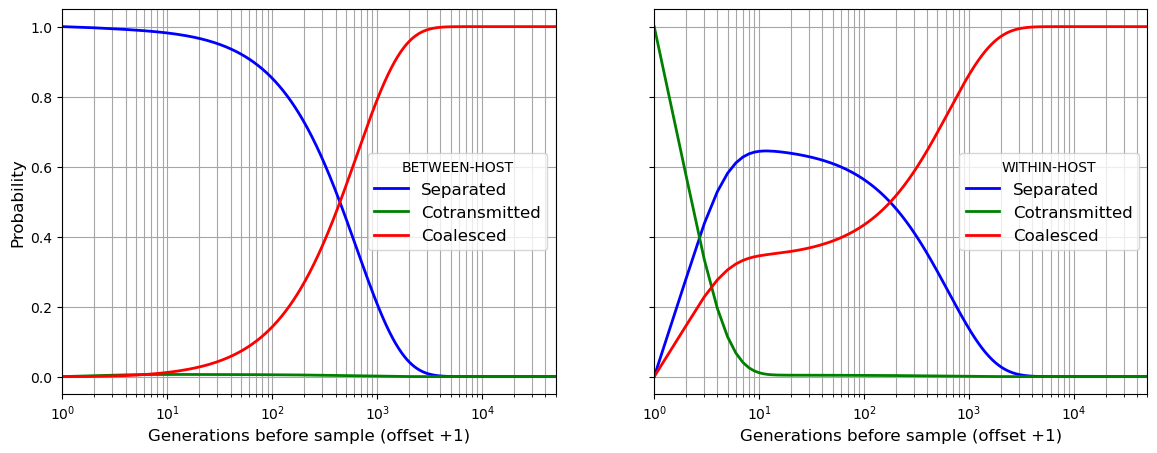
my_village.get_coalescent(show = True)
Observation time. Events captured. Mean coalescence time
beho wiho beho wiho
0 100.0 100.0 637.3 421.7
b_separated = my_village.beho_lineage[:, 0]
b_cotransmitted = my_village.beho_lineage[:, 1]
b_coalesced = my_village.beho_lineage[:, 2]
w_separated = my_village.wiho_lineage[:, 0]
w_cotransmitted = my_village.wiho_lineage[:, 1]
w_coalesced = my_village.wiho_lineage[:, 2]
# in this figure the time axis has a log scale
# .. which requires that we offset the axis by 1
# .. so we pretend that alleles are sampled at bt = 1
timescale = range(1, my_village.t_his + 1)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(14,5), sharex = True, sharey = True)
ax1.plot(timescale, b_separated, marker='', color='blue', linewidth = 2, label="Separated")
ax1.plot(timescale, b_cotransmitted, marker='', color='green', linewidth = 2, label="Cotransmitted")
ax1.plot(timescale, b_coalesced, marker='', color='red', linewidth = 2, label="Coalesced")
ax2.plot(timescale, w_separated, marker='', color='blue', linewidth = 2, label="Separated")
ax2.plot(timescale, w_cotransmitted, marker='', color='green', linewidth = 2, label="Cotransmitted")
ax2.plot(timescale, w_coalesced, marker='', color='red', linewidth = 2, label="Coalesced")
ax1.legend(title="BETWEEN-HOST",frameon=True, fontsize=12)
ax2.legend(title="WITHIN-HOST",frameon=True, fontsize=12)
ax1.set_xlabel("Generations before sample (offset +1)", fontsize=12)
ax2.set_xlabel("Generations before sample (offset +1)", fontsize=12)
ax1.set_ylabel("Probability", fontsize=12)
ax1.set_xscale('log',base=10)
ax1.set_xlim(1,my_village.t_his)
ax1.grid(visible=True, which='both', color='0.65', linestyle='-')
ax2.grid(visible=True, which='both', color='0.65', linestyle='-')
plt.show()
history_1 = [[10000, 100, 1, 0, 0]]
village_1 = cs.Population(history_1)
village_1.get_coalescent(show = True)
Observation time. Events captured. Mean coalescence time
beho wiho beho wiho
0 100.0 100.0 100.0 1.0
history_2 = [[10000, 100, 2, 0.5, 0]]
village_2 = cs.Population(history_2)
village_2.get_coalescent(show = True)
Observation time. Events captured. Mean coalescence time
beho wiho beho wiho
0 100.0 100.0 134.3 68.7
history_3 = [[10000, 100, 10, 1, 0]]
village_3 = cs.Population(history_3)
village_3.get_coalescent(show = True)
Observation time. Events captured. Mean coalescence time
beho wiho beho wiho
0 100.0 100.0 582.7 536.3
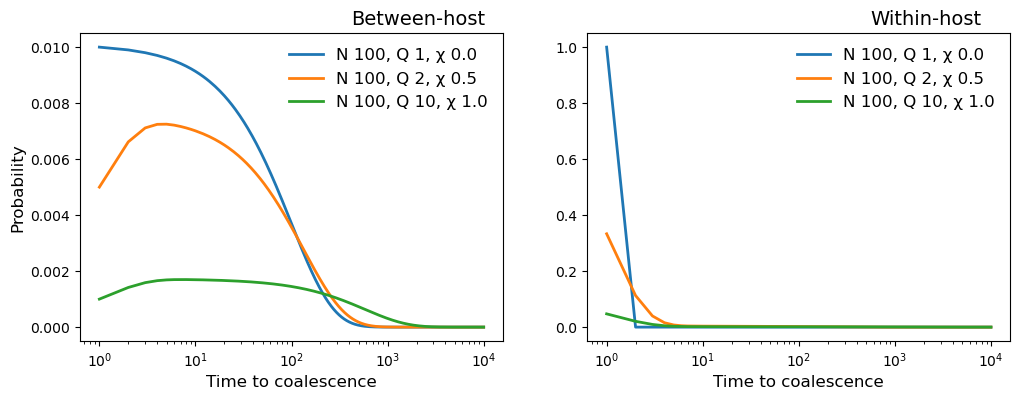
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12,4), sharex = True)
for example in [village_1, village_2, village_3]:
N = example.parameters[0,0]
Q = example.parameters[0,1]
X = example.parameters[0,2]
timescale = range(1, example.t_his)
beho_coalescent = example.coalescent[0, 1:, 0]
wiho_coalescent = example.coalescent[0, 1:, 1]
ax1.plot(timescale, beho_coalescent, marker='', linewidth=2, label=
"N {0:.0f}, Q {1:.0f}, \u03C7 {2:.1f}".format(N, Q, X))
ax2.plot(timescale, wiho_coalescent, marker='', linewidth=2, label=
"N {0:.0f}, Q {1:.0f}, \u03C7 {2:.1f}".format(N, Q, X))
ax1.set_title("Between-host", position=(0.8, 0.3), fontsize=14)
ax2.set_title("Within-host", position=(0.8, 0.3), fontsize=14)
ax1.set_xlabel("Time to coalescence", fontsize=12)
ax2.set_xlabel("Time to coalescence", fontsize=12)
ax1.set_ylabel("Probability", fontsize=12)
ax1.set_xscale('log',base=10)
ax1.legend(frameon=False, fontsize=12)
ax2.legend(frameon=False, fontsize=12)
plt.show()
view_past_generations = 5
print("Sample two alleles from different hosts\n")
print("Generations -------Probability distribution------")
print("back in time Separated Cotransmitted Coalesced")
for i in range(view_past_generations + 1):
separated = my_village.beho_lineage[i,0]
cotransmitted = my_village.beho_lineage[i,1]
coalesced = my_village.beho_lineage[i,2]
print("{0:6d}{1:16.2f}{2:13.2f}{3:14.2f}".format(
i,
separated,
cotransmitted,
coalesced ))
Sample two alleles from different hosts
Generations -------Probability distribution------
back in time Separated Cotransmitted Coalesced
0 1.00 0.00 0.00
1 1.00 0.00 0.00
2 0.99 0.00 0.00
3 0.99 0.01 0.00
4 0.99 0.01 0.00
5 0.99 0.01 0.01
print("Sample two alleles from the same host\n")
print("Generations -------Probability distribution------")
print("back in time Separated Cotransmitted Coalesced")
for i in range(view_past_generations + 1):
separated = my_village.wiho_lineage[i,0]
cotransmitted = my_village.wiho_lineage[i,1]
coalesced = my_village.wiho_lineage[i,2]
print("{0:6d}{1:16.2f}{2:13.2f}{3:14.2f}".format(
i,
separated,
cotransmitted,
coalesced ))
Sample two alleles from the same host
Generations -------Probability distribution------
back in time Separated Cotransmitted Coalesced
0 0.00 1.00 0.00
1 0.28 0.58 0.14
2 0.44 0.33 0.23
3 0.53 0.19 0.28
4 0.58 0.11 0.31
5 0.61 0.07 0.32

