Create a metapopulation#
We create a metapopulation in just the same way as creating a simple population by specifying its transmission history and invoking coalestr.Population(). It becomes labelled as a metapopulation when we create a subpopulation.
If we want to take account of historical patterns of parasite dispersal across the global metapopulation, a useful shortcut is to use the coalestr.species() function to construct a toy model of the emergence and expansion of Plasmodium falciparum as a species. The model has three phases:
a small founder population emerging approximately 15,000 years ago
an exponential growth phase beginning approximately 10,000 years ago
reaching a plateau of \(N_h \approx 15000\)
This gives a contemporary parasite population with a mean coalescence time of ~18,000 generations (assuming a generation time of 3 months) and a nucleotide diversity of \(\pi \approx 4 \times 10^{-4}\).
The model has default parameter settings which can be modified using keyword arguments as shown below.
Founder phase
founder_duration = 1000 (duration of the founder phase)
founder_N = 10
founder_Q = 3
founder_X = 0
Exponential growth phase
R0 = 1.0003
expansion_Q = 10
expansion_X = 0.1
Plateau phase
plateau_duration = 1000 (duration of the plateau phase)
plateau_N = 15000
plateau_Q = 10
plateau_X = 0.1
!pip install coalestr
from coalestr import cs
my_metapopulation = cs.species()
my_metapopulation.get_coalescent()
my_metapopulation.get_diversity()
Observation time. Events captured. Mean coalescence time
beho wiho beho wiho
0 100.0 100.0 17961.7 6421.2
Observation time. Nucleotide diversity Haplotype homozygosity
beho wiho beho wiho
0 3.95e-04 1.41e-04 3.23e-03 5.92e-01
Above we see that the default model gives a nucleotide diversity of 3.95e-4 in the current parasite population, and within-host nucleotide diversity of 1.41e-4.
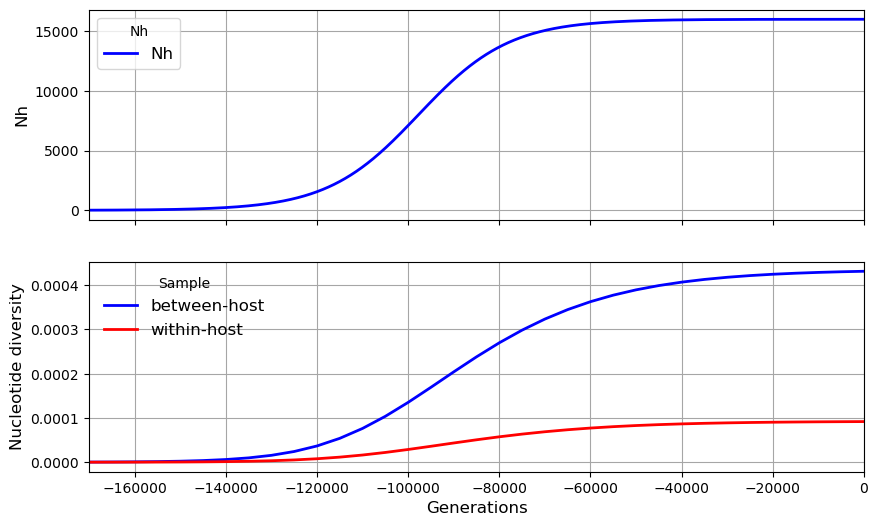
Let us look at time series data showing how nucleotide diversity builds up in the global metapopulation over time.
history = my_metapopulation.t_his
observe = [x for x in range(0,history,5000)]
my_metapopulation.get_coalescent(observe, show = False)
my_metapopulation.get_diversity(show = False)
my_metapopulation.plot_observations(metrics = ("Nh", "snp_het",))
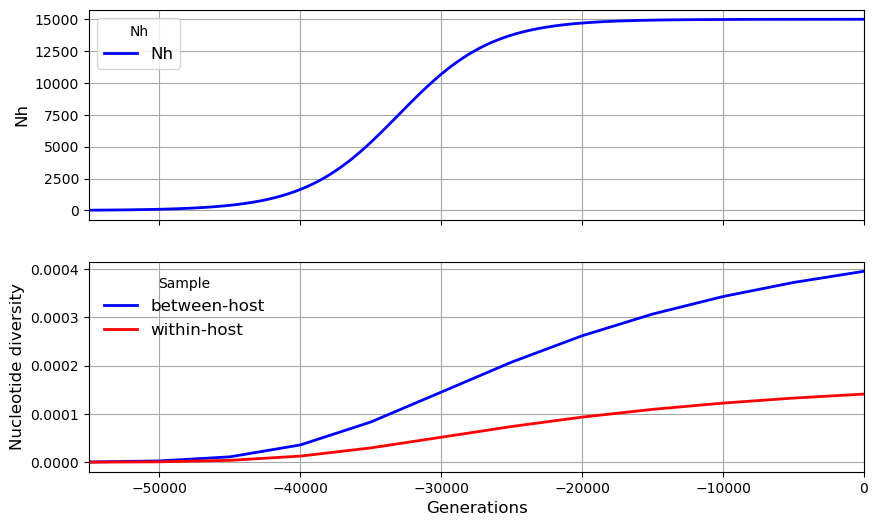
# Here we modify some of the default parameters
my_metapopulation = cs.species(
founder_duration = 1000,
founder_N = 10,
founder_Q = 3,
founder_X = 0,
R0 = 1.0001,
expansion_Q = 10,
expansion_X = 0.05,
plateau_duration = 1000,
plateau_N = 16000,
plateau_Q = 10,
plateau_X = 0.05)
history = my_metapopulation.t_his
observe = [x for x in range(0,history,5000)]
my_metapopulation.get_coalescent(observe, show = False)
my_metapopulation.get_diversity(show = False)
my_metapopulation.plot_observations(metrics = ("Nh", "snp_het",))