Nucleotide diversity#
We define nucleotide diversity (\(\pi\)) as the probability that two alleles are heterozygous at a random nucleotide position in the genome.
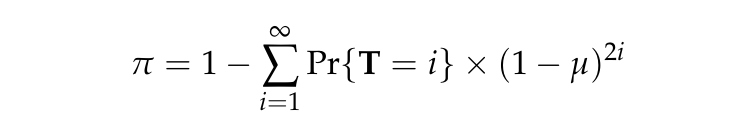
where T is a random variable representing time to coalescence and \(\mu\) is the single nucleotide substitution rate. From empirical observations we estimate that \(\mu = 1.1 \times 10^{-8}\) per generation.
We evaluate the above expression by using coalestr as follows:
We create a
Populationby specifying its transmission history.We use
get_coalescentto obtain the probability distribution of T.We use
get_diversityto evaluate the above summation series.
!pip install coalestr
from coalestr import cs
''' We create a history of transmission parameters
which in this case are constant over time
my_history = [[D, N, Q, X, M]]
D = duration of simulation
N = effective number of hosts (Nh)
Q = quantum of transmission
X = crossing rate of transmission chains (chi)
M = number of migrant hosts (Nm) '''
my_history = [[100000, 18764, 1, 0, 0]]
my_population = cs.Population(my_history)
my_population.get_coalescent()
my_population.get_diversity()
Observation time. Events captured. Mean coalescence time
beho wiho beho wiho
0 99.5 100.0 18188.4 1.0
Observation time. SNP heterozygosity. Haplotype homozygosity at 27.0 kb locus
beho wiho beho wiho
0 4.00e-04 2.20e-08 4.09e-03 9.87e-01
This tells us that for between-host (beho) comparisons, i.e. sampling from different hosts:
Our simulation captures 99.5% of coalescent events
Mean coalescence time is 18188.4 generations
Nucleotide diversity (aka SNP heterozygosity) is \(4 \times 10^{-4}\)
For within-host (wiho) comparisons, i.e. sampling from the same host:
Our simulation captures 100% of coalescent events
Mean coalescence time is 1 generation
Nucleotide diversity (aka SNP heterozygosity) is \(2.2 \times 10^{-8}\)
Haplotype homozygosity is discussed elsewhere.
In the following cells we will look at other combinations of transmission parameters that give us the same level of between-host nucleotide diversity, i.e. \(\pi = 4 \times 10^{-4}\) (which is approximately the level seen in the global parasite population).
def my_pop (D, N, Q, X, M):
pop = cs.Population(history = [[D, N, Q, X, M]])
pop.get_coalescent()
pop.get_diversity()
my_pop(100000, 18754, 10, 0, 0)
# N = 18754, Q = 10, X = 0
Observation time. Events captured. Mean coalescence time
beho wiho beho wiho
0 99.5 100.0 18188.8 10.0
Observation time. SNP heterozygosity. Haplotype homozygosity at 27.0 kb locus
beho wiho beho wiho
0 4.00e-04 2.20e-07 3.67e-03 8.85e-01
my_pop(100000, 18754, 1, 0.5, 0)
# N = 18754, Q = 1, X = 0.5
Observation time. Events captured. Mean coalescence time
beho wiho beho wiho
0 99.5 99.8 18180.0 9091.0
Observation time. SNP heterozygosity. Haplotype homozygosity at 27.0 kb locus
beho wiho beho wiho
0 4.00e-04 2.00e-04 4.09e-03 4.96e-01
my_pop(100000, 5568, 10, 0.5, 0)
# N = 5568, Q = 10, X = 0.5
Observation time. Events captured. Mean coalescence time
beho wiho beho wiho
0 99.5 99.6 18188.9 14213.0
Observation time. SNP heterozygosity. Haplotype homozygosity at 27.0 kb locus
beho wiho beho wiho
0 4.00e-04 3.13e-04 3.99e-03 2.14e-01
my_pop(100000, 3269, 10, 1, 0)
# N = 3269, Q = 10, X = 1
Observation time. Events captured. Mean coalescence time
beho wiho beho wiho
0 99.5 99.6 18187.4 16687.4
Observation time. SNP heterozygosity. Haplotype homozygosity at 27.0 kb locus
beho wiho beho wiho
0 4.00e-04 3.67e-04 4.05e-03 8.44e-02

